- Tools
- Downloads
- Links
- Community
- Species
- About
- Help
FB2024_02
,
released April 23, 2024
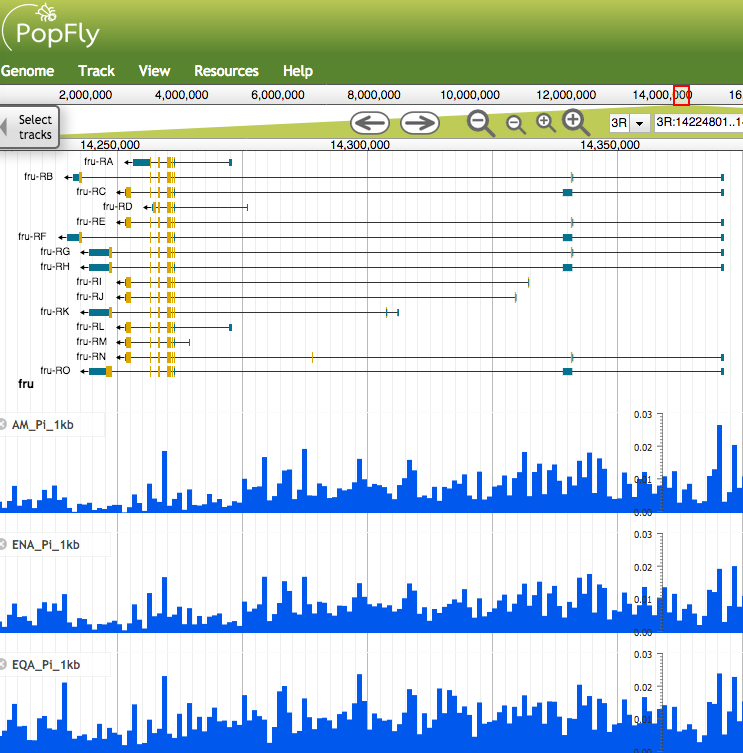
With release FB2017_05, FlyBase introduces links from FlyBase Gene Reports to the PopFly Drosophila Population Genomics Browser, produced by colleagues in the Bioinformatics of Genome Diversity group from the Universitat Autònoma de Barcelona (UAB) and the Institut de Biotecnologia i Biomedicina (IBB) (Hervas S, Sanz E, Casillas S, Pool JE, and Barbadilla A (2017) PopFly: the Drosophila population genomics browser. Bioinformatics, 33, 2779-2780; https://doi.org/10.1093/bioinformatics/btx301).
The PopFly browser provides sophisticated visualization and download of nucleotide diversity metrics, linkage disequilibrium statistics, recombination rates, neutrality tests, and population differentiation parameters at non-overlapping windows of varying size from the over 1100 Drosophila melanogaster genome sequences compiled in the Drosophila Genome Nexus data-set (Lack JB, Lange JD, Tang AD, Cortett-Detig RB, Pool JE (2016) A Thousand Fly Genomes: An Expanded Drosophila Genome Nexus. Mol. Biol. Evol. 2016 Dec; 33(12): 3308-3313).
For simplicity's sake, links from FlyBase Gene Reports are configured to display only Pi nucleotide diversity statistics for the six geographical Drosophila "metapopulations" compiled by the PopFly group (America, Asia, Equatorial Africa, Europe / North Africa, Oceania, and Southern Africa). In order to take advantage of the rich population genomics data available from PopFly, we encourage new users to consult the excellent documentation and PopFly Tutorial available under the Help menu on the PopFly site.