- Tools
- Downloads
- Links
- Community
- Species
- About
- Help
FB2024_02
,
released April 23, 2024
FlyBase will list here changes and improvements to the website, organized according to FlyBase releases, with the most recent release at the top.
Subscribe to the FlyBase Substack
to stay up-to-date with the latest news from FlyBase including release announcements,
web site updates, and important Drosophila community news.
This is a low volume newsletter.
 Newest Tools link
Newest Tools link
Transgenic alleles now have a “Transgenic product class” field that uses Sequence Ontology (SO) term(s) to describe the nature of the encoded product, so that it is easier for you to see whether it is useful for your experiments. The transgenic product class terms we use fall into 4 broad types:
The field is located in the General Information and Nature of the Allele sections of the Allele Reports.
To get a list of all transgenes that encode a particular type of gene product, either click on the SO term in the “Transgenic product class” field of an individual Allele Report, or use the Vocabularies tool to go to the relevant Term Report. On the Term Report, click on the Alleles button in the Annotations section to get a hist list of all the matching transgenic alleles. For example, the wild_type term lists over 28,000 transgenic alleles, while missense_variant lists almost 7800 alleles.
FlyBase has updated the DRSC Integrative Ortholog Prediction Tool (DIOPT) to version 9.1. This update is derived from the latest data available from the DRSC's DIOPT tool and includes the following orthology prediction algorithms: Compara, eggNOG, Hieranoid, Homologene, InParanoid, OMA, OrthoDB, OrthoFinder, OrthoInspector, OrthoMCL, Panther, Phylome, SonicParanoid, and Domainoid.
DIOPT 9.1 now also includes predictions for A. gambiae (African malaria mosquito) and E. coli.
As with previous versions, DIOPT 9.1 predictions can be accessed from the FlyBase home page using the “Homologs” search tool, or viewed on gene reports (“Orthologs” and “Paralogs” sections).
For further details regarding DIOPT and its features, please visit the DRSC's DIOPT website.
See the 2021_06 Release Notes for details of changes to the FlyBase database for this release.
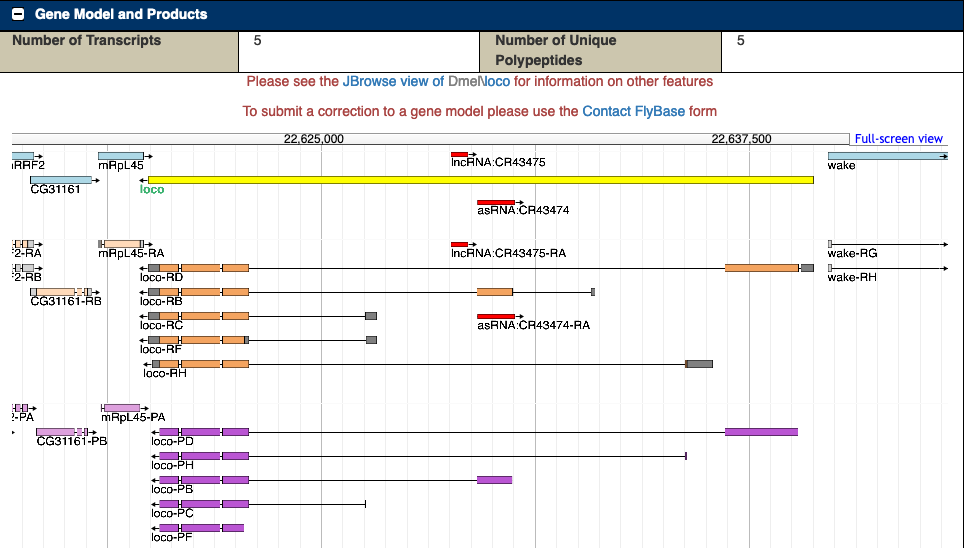
FlyBase now reflects gene model annotation and genome assembly information for D. melanogaster only. As of this release (FB2020_03), annotation and assembly information for these species has been retired: D. simulans, D. ananassae, D. pseudoobscura and D. virilis. This follows retirement of such information in release FB2018_06 for the following species: D. erecta, D. grimshawi, D. mojavensis, D. persimilis, D. sechellia, D. willistoni, D. yakuba. For the status of these gene model annotations, please see the NCBI page Eukaryotic genomes annotated at NCBI.
The FlyBase BLAST tool will continue to support queries against the reference genomes of these retired species, but not queries against annotated transcripts or proteins. For the current and future releases, there will be no JBrowse or GBrowse view of the gene model annotations for the species. FASTA and GFF files for the retired gene model annotations and assemblies will still be available on the FlyBase FTP site ftp://ftp.flybase.net/genomes/, but they will not be updated. The related gene reports will persist, containing various types of curated information (phenotypes, expression, publications), as well as links to sequence information at external sites like NCBI Gene in the "CrossReferences" section, though these links may go stale over time.
We emphasize that this decision was carefully deliberated, made in consultation with the FlyBase advisory board and our partners at the NCBI, to allow FlyBase to redirect its energies from these rarely used resources to other aspects of the project that are more heavily used by the Drosophila community. Please see this NCBI Announcement to learn more about their Drosophila gene annotation resources.
Our ID Converter tool is now called ID Validator. This tool will accept a list of FlyBase symbols/IDs (for any data type) and, where necessary/possible, update them to their current versions. It will also convert certain external IDs (GenBank nucleotide/protein accessions, UniProt accessions, PubMed IDs) into their equivalent FlyBase IDs. The output is provided as a validation table that can either be downloaded as a file or exported to a FlyBase HitList for futher processing (including conversion between data types).
Changes include:genetically encoded sensor Term Report.
SRA aggregated RNA-Seq (Oliver Lab) tracks in the “Expression Levels: RNA-Seq” section;
tracks are offered with signal cut-off set to 100 (high sensitivity), 1,000 (medium sensitivity) or 10,000 (low sensitivity).
proteomic peptides (uniquely mapping) track,
listed in the “Aligned evidence” section of JBrowse tracks.
Peptides from additional proteomic studies will be added to this track over time.
Tools → People in the FlyBase page navigation bar.
You can search for other Drosophila researchers, or you can add or update your contact information in our database.
*Polytene images were first made available in October 2016.
**A full map correspondence table of recombination,
cytology and sequence map values for D. melanogaster genes on the reference genome assembly is available in the
Maps section of the FlyBase Resources page.
*Recombination map data were first made available in FB2016_05 (September 2016).
**The new "Recombination map" section was first released in FB2017_01 (February 2017).
Protein domains (PFAM) under the
"Aligned Evidence" section of the tracks listing)
showing protein domains, where you can see these domains aligned
with the genome sequence and protein transcript tracks.
RAMPAGE TSS or MachiBase tracks
listed under the "Aligned Evidence" section of the track selection page.
Developmental stages, stranded small RNA-Seq (Lai lab)
Tissues, stranded small RNA-Seq (Lai lab)
Tissue culture cells (Schneider + embryonic), stranded small RNA-Seq (Lai lab)
Tissue culture cells (imaginal disc), stranded small RNA-Seq (Lai lab)
Tissue culture cells (CNS, ovary, blood), stranded small RNA-Seq (Lai lab)
Links → External Resources.
Help → Video Tutorials.
Histone Modifications - mesoderm (Furlong lab, ChiP-Seq peak calls)
TFBS - mesoderm (Furlong lab, ChiP-chip)
Tools ⇒ RNA-Seq Tools),
and the QuickSearch Expression tab now has a panel with links to each tool.
Each of these access points also has a link to a new
RNA-Seq tools summary page,
where you can learn more about how and when to use each of these tools,
with links to the tools and in-depth help.