- Tools
- Downloads
- Links
- Community
- Species
- About
- Help
FB2024_02
,
released April 23, 2024
The “GAL4 etc” QuickSearch tab has been redesigned to allow you to search a wider selection of transgenic driver and reporter alleles. In the original GAL4 etc tab, you could search for only three binary drivers (GAL4, QF, and lexA), and two reporters (lacZ and GFP). With the incorporation of Experimental Tools, you now have access to all the ‘et cetera’ of GAL4 etc.
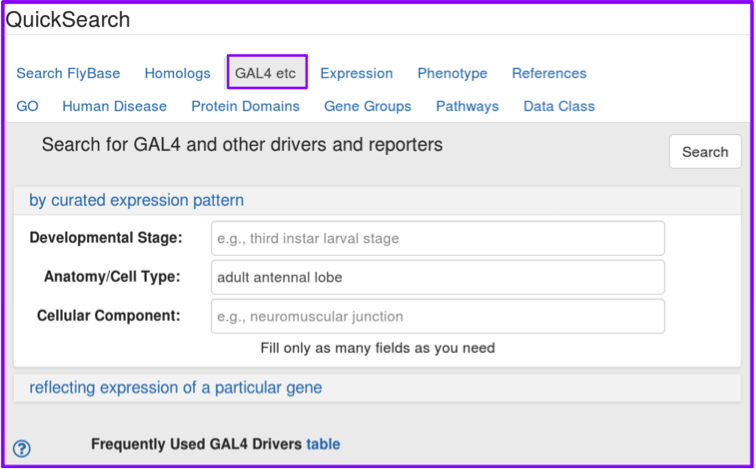
The first change you may notice in the redesigned GAL4 etc tab is the absence of the Driver/Reporter pull-down menu: you will be able to filter for the driver/reporter you want in a more robust and detailed manner on the results page. As with the original GAL4 etc tab, you can search for curated expression patterns by developmental stage, anatomy or cell type, and/or cellular component, using valid FlyBase Development and Anatomy controlled vocabulary terms, and GO cellular component terms. We’ve added a new search option: you can now search for drivers/reporters that have been curated as reflecting the expression pattern of a particular gene, such as dpp.
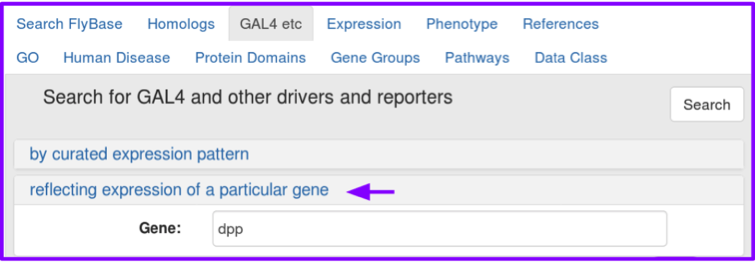
These search options are mutually exclusive: you can search by curated expression pattern or by gene, but not by both at once.
The hitlist resulting from either search includes all drivers and reporters. For some searches the hitlist may be very large, as in this search for the term “adult antennal lobe”, which has over 4600 results.
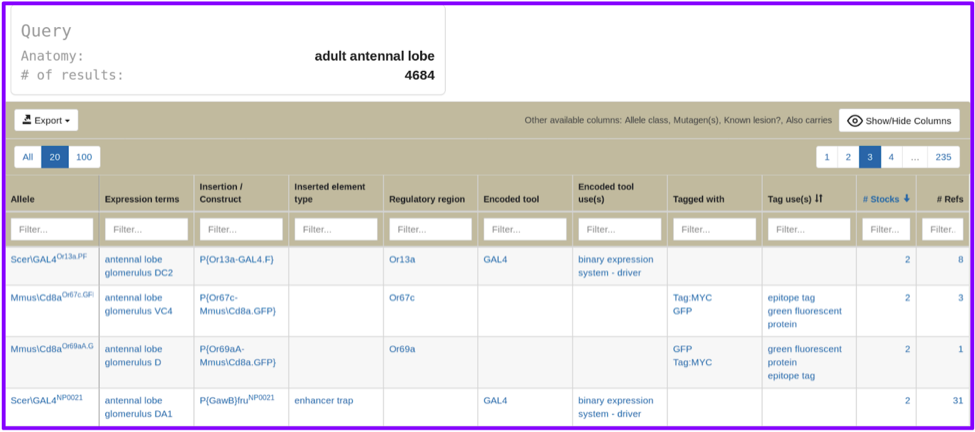
This hitlist has been designed to allow you to easily find the results you’re looking for, using the same features now incorporated into the Alleles, Insertions, and Transgenic Constructs section of gene reports:
Each column can be filtered using the text box at the top of each column. For example, if you’re looking for a set of lexA drivers expressed in an adult antennal lobe, you can filter for that by entering ‘lexA’ in the Encoded Tool box. This filtered search results in a much more constrained hitlist, in this case, fewer than ten results. (Note that some columns have been closed).
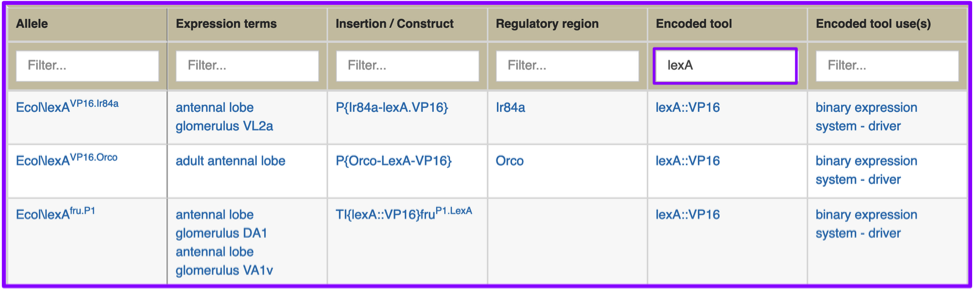
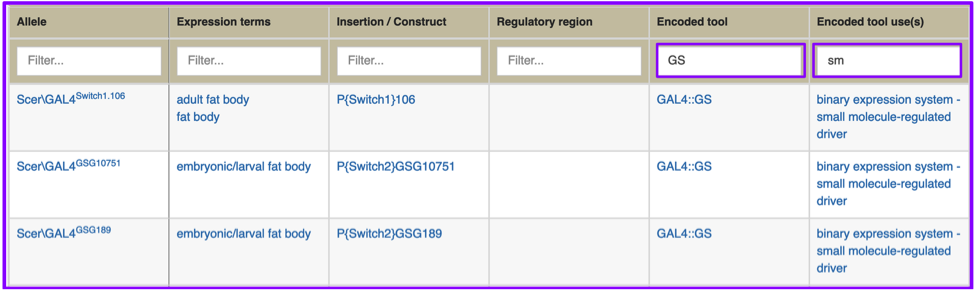
You don’t need to know the exact tool symbol or controlled vocabulary term to search for a tool or class of tool to find what you’re looking for; column filtering is text-based, and will restrict as you type. You may, for example, want a list of fat body GAL4 drivers that use the mifepristone-inducible GeneSwitch tool; searching specifically for GeneSwitch GAL4 drivers was not an option with the previous GAL4 etc tab. After doing a search for “fat body”, you can enter ‘GS’ in the Encoded tool box, finding all instances of GAL4::GS, or ‘sm’ into Encoded tool use(s), finding all instances of binary expression system - small molecule-regulated driver.
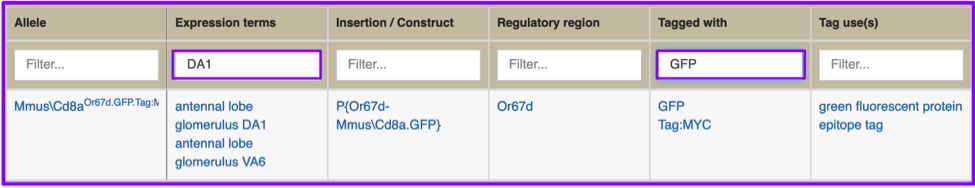
You can also filter multiple columns at once. In this example, the “adult antennal lobe” hitlist has filtered to show only proteins tagged with GFP (filtered using Tagged with box) that are expressed in antennal lobe glomerulus DA1 (filtered using the Expression terms box). Note that controlled vocabulary terms are now hyperlinked to the relevant Vocabularies term report.
Note that if you realize you’ve picked too general an anatomy term (over 4500 GAL4 drivers are expressed in the adult antennal lobe), you can use the Expression terms column, which includes every anatomy term with curated expression data that has a part_of or is_a relationship to the term you searched with, allowing you filter with a more specific controlled vocabulary term, rather than starting a new search.
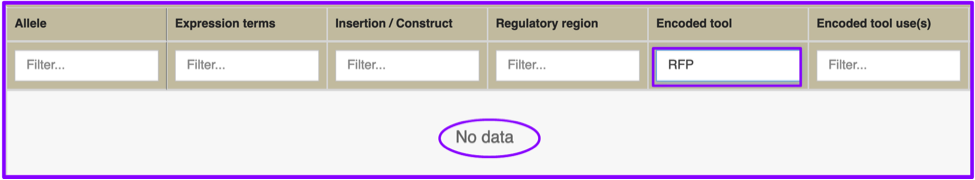
The beginning of this commentary promised the ability to search for drivers/reporters that could not be found by the previous GAL4 etc tab. The final example is of an ‘et cetera’: promoter fusion reporter that expresses red fluorescent protein the adult antennal lobe. Your initial attempt, filtering the Encoded tool column with the text string “RFP” fails, but you’re certain you’ve seen examples of that expression pattern in papers.
Instead, you try typing “fluorescent” into Encoded tool use(s). This results in a hitlist that includes two red fluorescent protein reporters, one of which is the tool DsRed(Unk), and the other is mCherry; this filter also found two EGFP reporters.
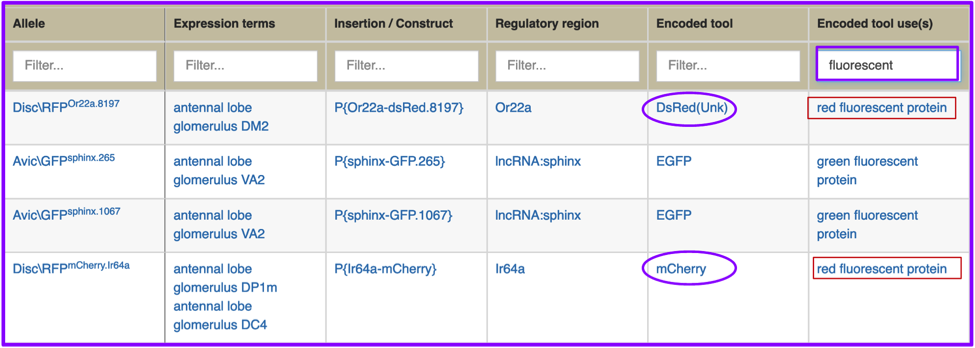
You can also export your results to a standard FlyBase hitlist, a comma-separated file, or an Excel file, using the Export button. The filters you’ve set in your hitlist will be exactly maintained in your exported hitlist or file; exporting the final example will result in a hitlist or file containing these four alleles, not the nearly 4700 alleles in the initial unfiltered hitlist.