- Tools
- Downloads
- Links
- Community
- Species
- About
- Help
FB2024_02
,
released April 23, 2024
Around 30% of protein-coding genes in D. melanogaster encode enzymes, and at least 15% of the genome encodes factors involved in core metabolic pathways. Relevant FlyBase Gene Reports now show improved information on enzyme nomenclature and reactions, as well as links to metabolic pathway resources, which we hope will support the growing number of researchers working in these fields.
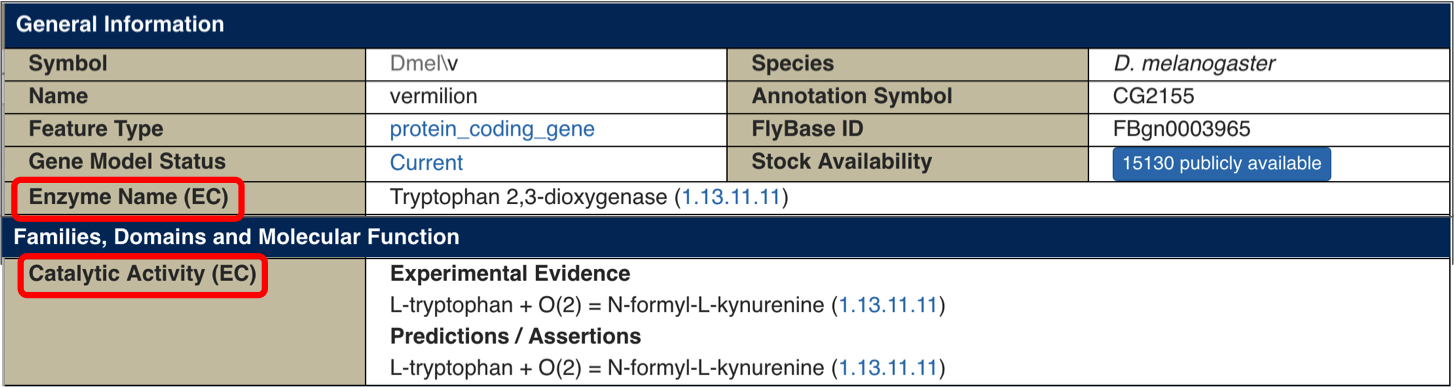
The uppermost ‘General Information’ panel on Gene Reports now contains an ‘Enzyme Name (EC)’ field that displays the systematic name for a given enzyme together with its Enzyme Commission (EC) number. (Showing the systematic name is especially useful where the fly gene is either unnamed or has been named based on its mutant phenotype rather than its wild type function.) Further down the page, in the ‘Families, Domains and Molecular Function’ section, the new ’Catalytic activity (EC)’ field displays the reaction(s) catalyzed by the enzyme. The EC data in both these new fields are derived from our Gene Ontology (GO) Molecular Function annotations, taking advantage of the EC cross-references within the GO itself. This allows the catalytic activities to be separated into those based on experimental evidence versus those based only on predictions or computational assertions, using the evidence code associated with the underlying GO annotations.
We have also introduced a new ‘Pathways’ section within the Gene Report. Where relevant, this section now includes the names of metabolic (and other) pathways in which the gene product is involved, with links to the respective reports at KEGG and Reactome. Additional links to BioCyc are due in the near future. This section also includes a link to and description of our growing collection of FlyBase-curated reports for signalling pathways, where relevant.

TCA Cycle illustration by Tamara Clark, www.tamaraclark.com.