- Tools
- Downloads
- Links
- Community
- Species
- About
- Help
FB2024_02_2
,
released April 23, 2024
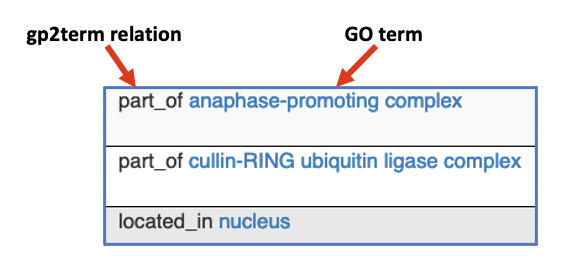
FlyBase is introducing gene product to term (gp2term) relations for Gene Ontology (GO) annotations. These new relations are aimed at giving more contextual information to the GO term used. For example, lmgA is annotated with the GO terms ‘anaphase-promoting complex’ and ‘nucleus’. These will now be displayed with gp2term relations to read: ‘part_of anaphase-promoting complex’ and ‘located_in nucleus’.
For this release, a limited, default set of gp2term relations will be visible in the ‘Function’ section of gene reports: ‘enables’ or ‘contributes_to’ for molecular function, ‘involved_in’ for biological process, and, for the cellular component aspect of the GO, ‘part_of’ for protein complexes, ‘located_in’ for cellular compartments and ‘colocalizes_with’ for localization which is proximal to a component. In the next release, we will start to add annotations with more granular gp2term relations and, over time, this change will improve the accuracy of GO annotations and bring them into line with the current plans of the GO consortium.
These changes will be reflected in the Gene Association File (GAF), which will be updated to the GAF2.2 specifications. For this release, we will provide the standard GAF as the default download and provide GAF2.2 for those users that wish to switch to this format or test parsing software. GAF2.2 will become the default GAF for FB2021_01, but the old format will be maintained for a period of time to give users time to accommodate the changes.