- Tools
- Downloads
- Links
- Community
- Species
- About
- Help
FB2024_02
,
released April 23, 2024
We have added new analysis of genomic variants data to our Gene reports, Allele reports, and JBrowse. This represents a collaboration between FlyBase, the Alliance of Genome Resources, and Ensembl. Variants data from FlyBase and other participating model organisms are run through the Ensembl VEP (Variant Effect Predictor) pipeline as a collaboration between the Alliance and Ensembl, and the results are currently displayed at the Alliance. With this release, the Drosophila results of this pipeline are also available on FlyBase.
The VEP algorithm considers the genomic location, mutation type, and nucleotide change of a variant to calculate the molecular consequence on all of the overlapping transcripts affected by the variant. In addition to reporting the molecular change to the transcripts and encoded CDS, it assigns a Sequence Ontology (SO)-based consequence term to each variant/transcript combination. A glossary of terms used by the VEP can be found here.
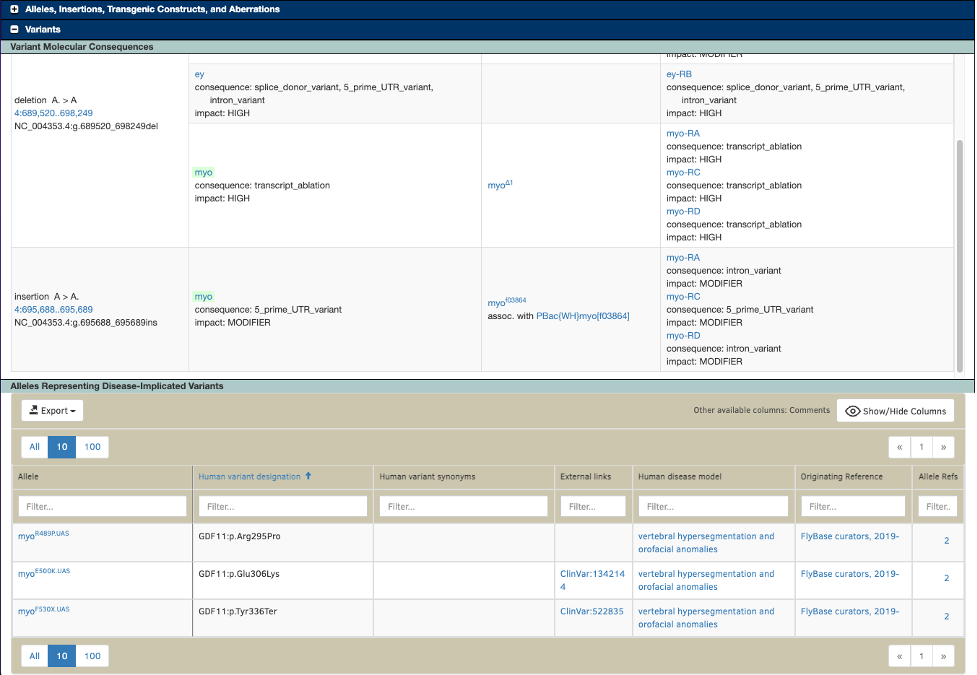
The VEP results are displayed in FlyBase in a new “Variants” section of the Gene Report, under the subsection “Variant Molecular Consequences”. All of the variants associated with a gene that have been run through the VEP pipeline appear in a scrolling table. The table displays, for each variant, all genes that are potentially affected by that variant, any alleles of those genes associated with the variant, and the predicted consequences to all affected transcripts. The gene that is the subject of the report is highlighted in the table. See for example myo.
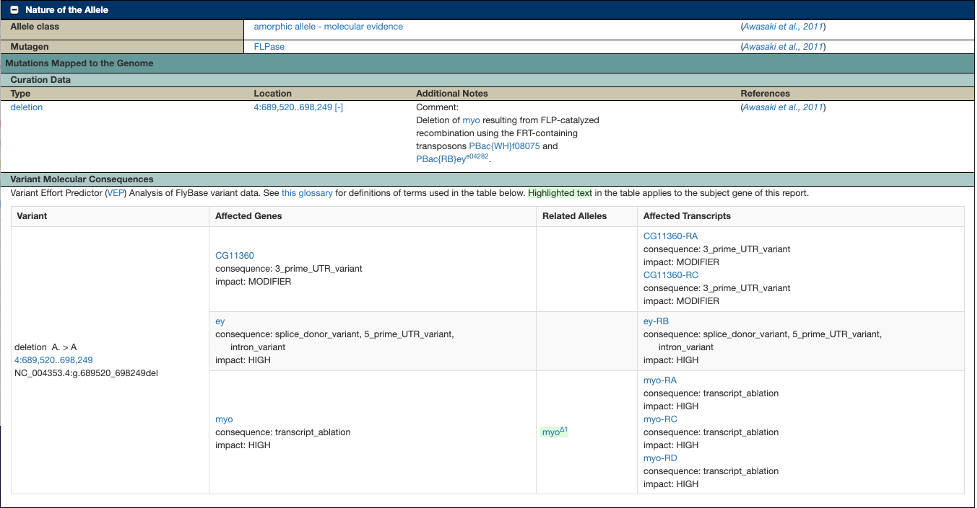
VEP analysis results are also available from individual allele reports. The new “Variant Molecular Consequences” section appears under the FlyBase description of the variant found in the “Mutations Mapped to the Genome” subsection, under “Nature of the Allele”.
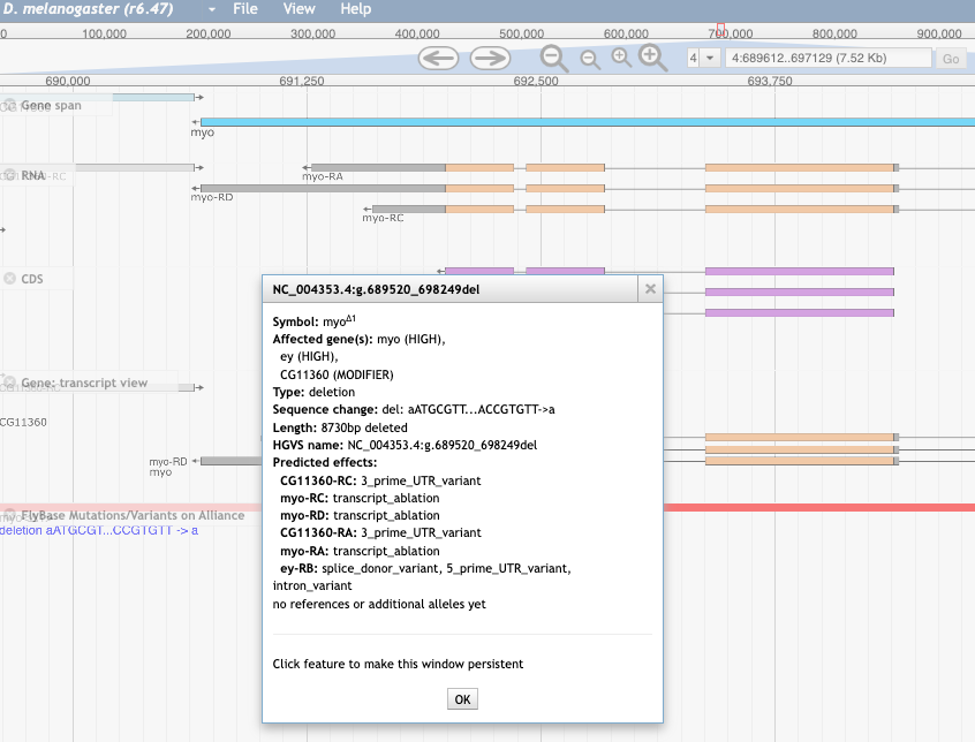
Finally, the variants and VEP analysis results can be seen on a new “FlyBase Mutations/Variants on Alliance” track that can be found in the “Mutations & Sequence Variants” section of JBrowse. If you click on a mutation, a popup box appears containing the VEP analysis data.